The composite format allows including all types of blood and tissue data into a single tab-delimited text file. The advantage is that all data can be prepared externally and loaded at once, and that such files can directly be used for batch processing.
File Start
The file starts with the two lines

It is followed by a sequence of sections for the data and other related information.
Demographic and Study Data
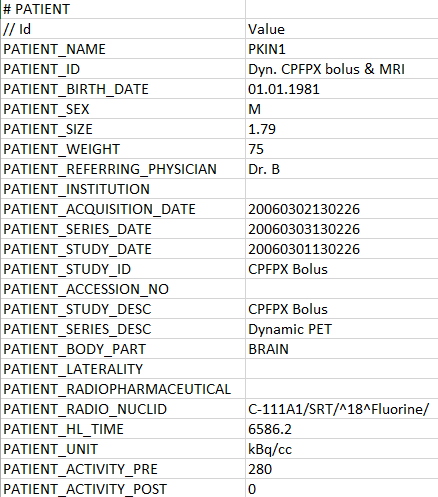
The demographic information and study-related data can be defined in a section headed by the # PATIENT tag. Each line starts with a tag in capital letters which is followed by a tab, and then the corresponding value.
Comments
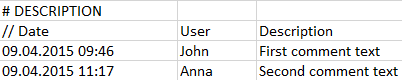
Comments can be included for describing the data and recording important facts. The # DESCRIPTION section allows specifying text which will be imported with the data and can be shown in the user interface. The three elements to enter in a line are the Date of the comment, the User who edited it, and the actual comment text as the Description.
Further comments can be added as lines preceded by a // in a # NON_PMOD_COMMENTS section. They are only for documentation purposes of the raw data and not interpreted by PKIN.

Model Specification
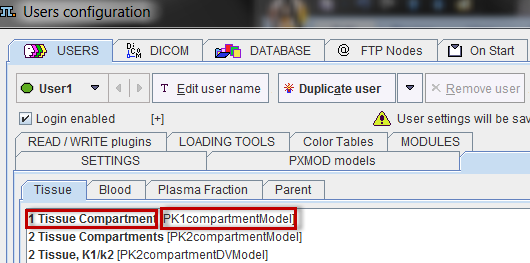
A tissue model can optionally be specified in a # MODELS section.

The Model entry corresponds to the class name, and the Name entry to the string shown in the model list. Both can be determined in the configuration utility as illustrated below.
For reference models the reference region can be specified in by an additional line with the REFERENCE_REGION tag.

Note that the region number (starting from 0) is relevant, not the region name.
Data Sections
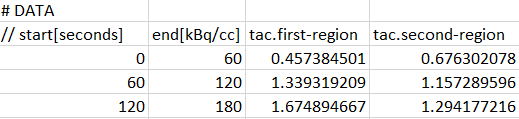
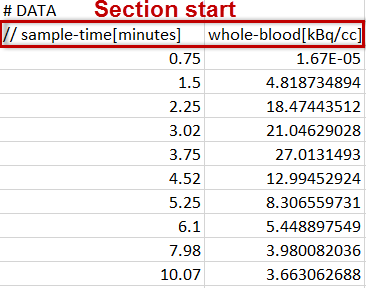
Each data part to be included is specified in a section starting with a line # DATA followed by a header line of the actual data. For blood data the following format is supported. Note the data units which are specified in the brackets directly after the column names in the header line.
For tissue data, it is assumed that the first two columns define the acquisition start and end times, whereas the following columns contain the data for the different tissue regions.
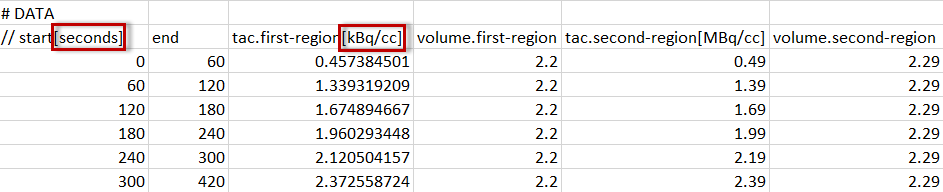
Each region is represented by a column with header tac.name, where "name" will be shown as the region name in PKIN. Two regions will result from the example above, with names first-region and second-region. Again, the data units are specified in brackets, directly after the region names. Optionally, the volumes corresponding to the TAC can be defined in additional columns, indicated by the header volume.name. The volume unit is cc, so there is no need to specify it.
Multiple blood and tissue data sections can be included in a composite data file, for instance to handle TACs with different timing. The following key words are parsed in the header lines.
sample-time |
Timing of blood samples. |
whole-blood |
Activity of whole blood samples, to be used for spillover correction. May also be used for calculating the plasma activity if a plasma fraction is loaded. |
plasma.1 |
Plasma activity of the first input curve, the second, etc. Plasma activity may represent the activity of parent, if the metabolite correction has been done outside of PKIN, otherwise the activity of parent and metabolites. In the latter case a parent fraction has to be loaded for the metabolite correction within PKIN. |
plasma-fraction.1 |
Plasma fraction of the first input curve, the second, etc. These fractions will be multiplied with whole-blood for the calculation of plasma.1, plasma.2 etc. Supported units: [1/1], [%]. |
parent-fraction.1 |
Parent fraction of the first input curve, the second, etc. These fractions will be multiplied with plasma.1, plasma.2 etc. for the calculation of the input curves. Supported units: [1/1], [%]. |
start |
Start time of the PET/SPECT acquisition frames. The time units of the tissue TACs are specified after begin, eg. end[seconds]. |
end |
End time of the PET/SPECT acquisition frames. |
tac.name |
Tissue TAC column with the name specified after the tac. tag. |
volume.name |
Volume of the tissue TAC column with the name specified after the volume. tag. Unit of the volume is cc. |
tac.reference |
Tissue TACs with this label will be used as reference when applying reference tissue models, if REFERENCE_REGION is not specified in the model section. |
File Format and Loading
The information described above can be saved in a tab-delimited text file (.kmData) or an XLS (Excel 2003, not xlsx) file and loaded with the Load KM File entry from the Kinetic menu. Alternatively, the file can be dragged and dropped onto the PKIN window or the PMOD ToolBox.