Calibration Measurement
A calibration measurement should be performed as described above, measuring the response in the blood counter to a rectangular input activity. Note that the measured data have to be decay corrected and calibrated to kBq/cc.
Fitting of the Dispersion Parameters
In PKIN, please start a new workspace and load the measurement with the menu item Load Whole Blood Activity. On the Blood tab, switch the Whole Blood model to Calibration: Step-up as illustrated below.
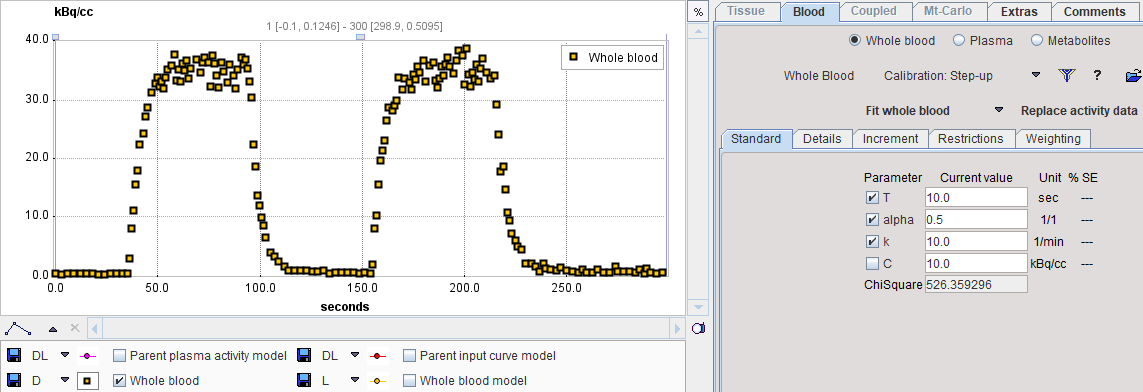
Mask all samples except for a baseline, rising edge and the following plateau as follows:
1.Right click into the curve area. In the context menu
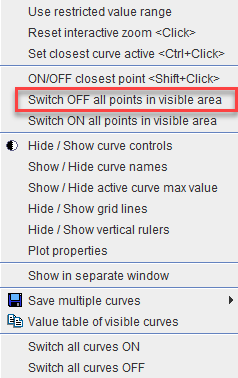
select Switch OFF all Points in visible area. Consequently all samples appear gray, meaning they are not considered in fitting operations.
2.Open the context menu again and select Use restricted value range. Enter the start/end times of the relevant section and close with Ok.
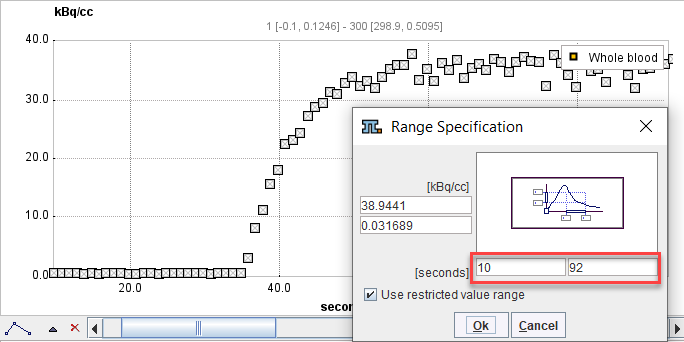
3.Open the context menu again and select Switch ON all Points in visible area.
Next comes the fitting of the step-up model. Enter the time where the signal rises as a starting value for the T parameter, the known concentration value as the C parameter, and enable the fitting flags as illustrated below.
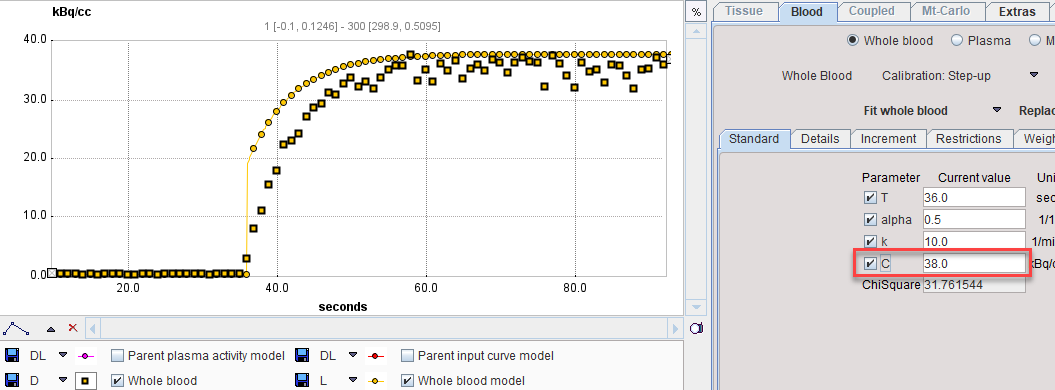
With Fit Whole Blood the model curve can be fitted to the section.
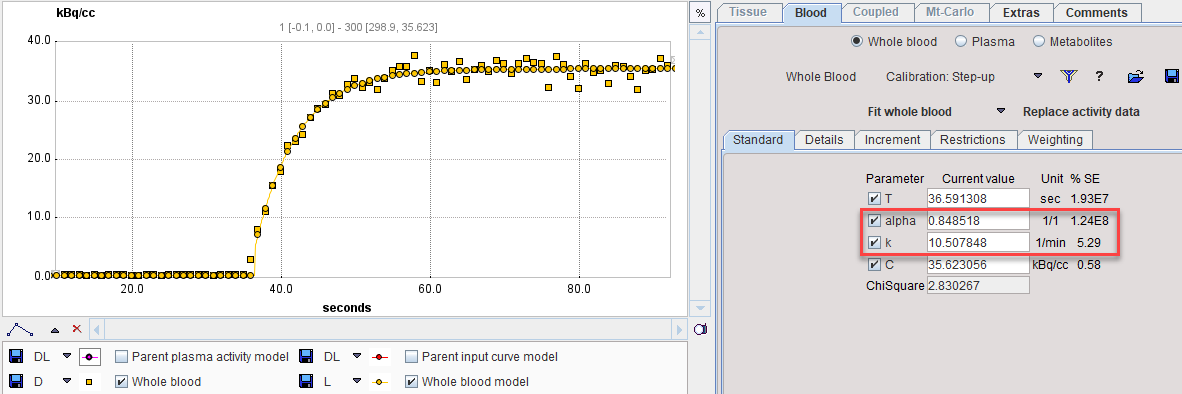
A similar fitting can be done with the decreasing edge and the model Correction: Step-down. Fitting the different parts of the calibration experiment provides several sets of the dispersion parameters, which allows determining their average values. Those values of the alpha and k parameters can be applied for the correction of the actual blood curve of live experiments.
Recommendations by Methodology Author, Ole Munk
alpha has an impact on the noise in the dispersion-corrected input functions. If alpha is allowed to get close to 1, then the noise increases (too) much (Fig.8 in Munk et al.). Therefore, in practice, alpha can be fixed at a reasonable value, eg. 0.75, and only k fitted along with the two "known" parameters T and C.
Notes:
1.The parameters can be tracer-dependent. It takes less than an hour to do the calibration measurement and the parameters can be re-used for all future measurements using the same blood sampling setup.
2.Dispersion correction is only important when modeling fast processes such as blood flow. When estimating binding potentials and other "slow processes'", dispersion correction is considered less important.