uNET is a convolutional neural network developed for image segmentation (Ronneberger et al., 2015).
A modified uNET was used by Isensee et al. (2018) in the MICCAI Brain Tumor Segmentation Challenge (BraTS). The BraTS data and this modified uNET were used for our first case study and the Multichannel Segmentation architecture for segmentation in PAI. The modified uNET structure is illustrated here:
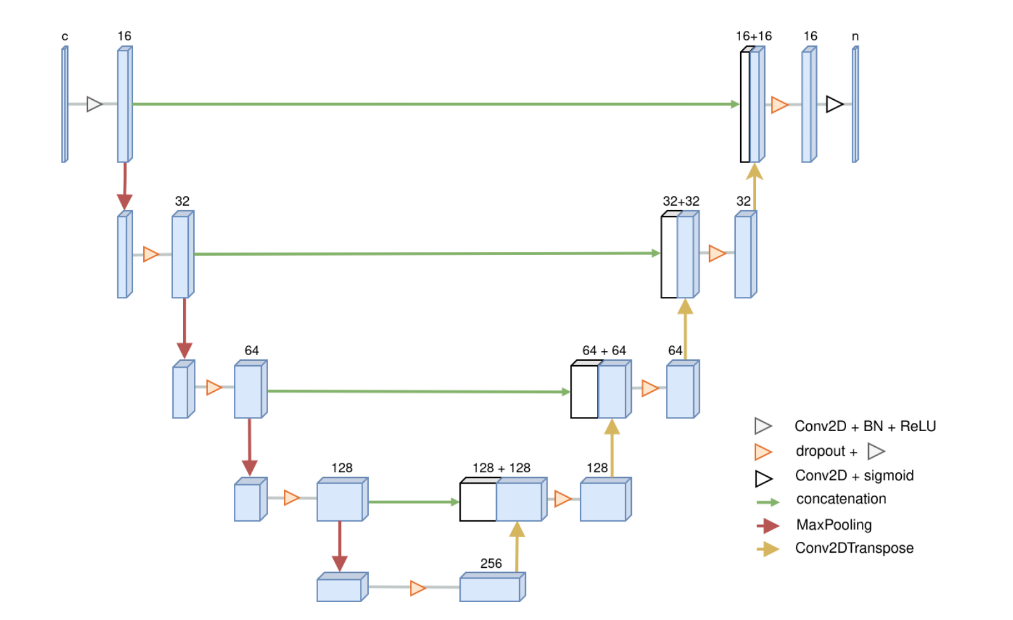
Additional case studies were made using this architecture:
•Rat brain dopaminergic PET segmentation
•Human brain MRI deep nuclei segmentation (IXI)
•Human cardiac MRI segmentation for function analysis
This architecture is available as Multichannel Segmentation in the Create Learning Set dialog.
The loss function used for this architecture is Dice Coefficient.
A classical uNET architecture is also available for segmentation projects. It is available as uNET Segmentation in the Create Learning Set dialog. Its structure for 2D input data is illustrated here:

This architecture is used for mouse cardiac MRI segmentation in the PCARDM tool, and it has additionally been used for the following case study:
•mouse femur/tibia trabecular segmentation
The loss function used for this architecture is Binary Cross Entropy.
For 3D training data its structure is illustrated here:
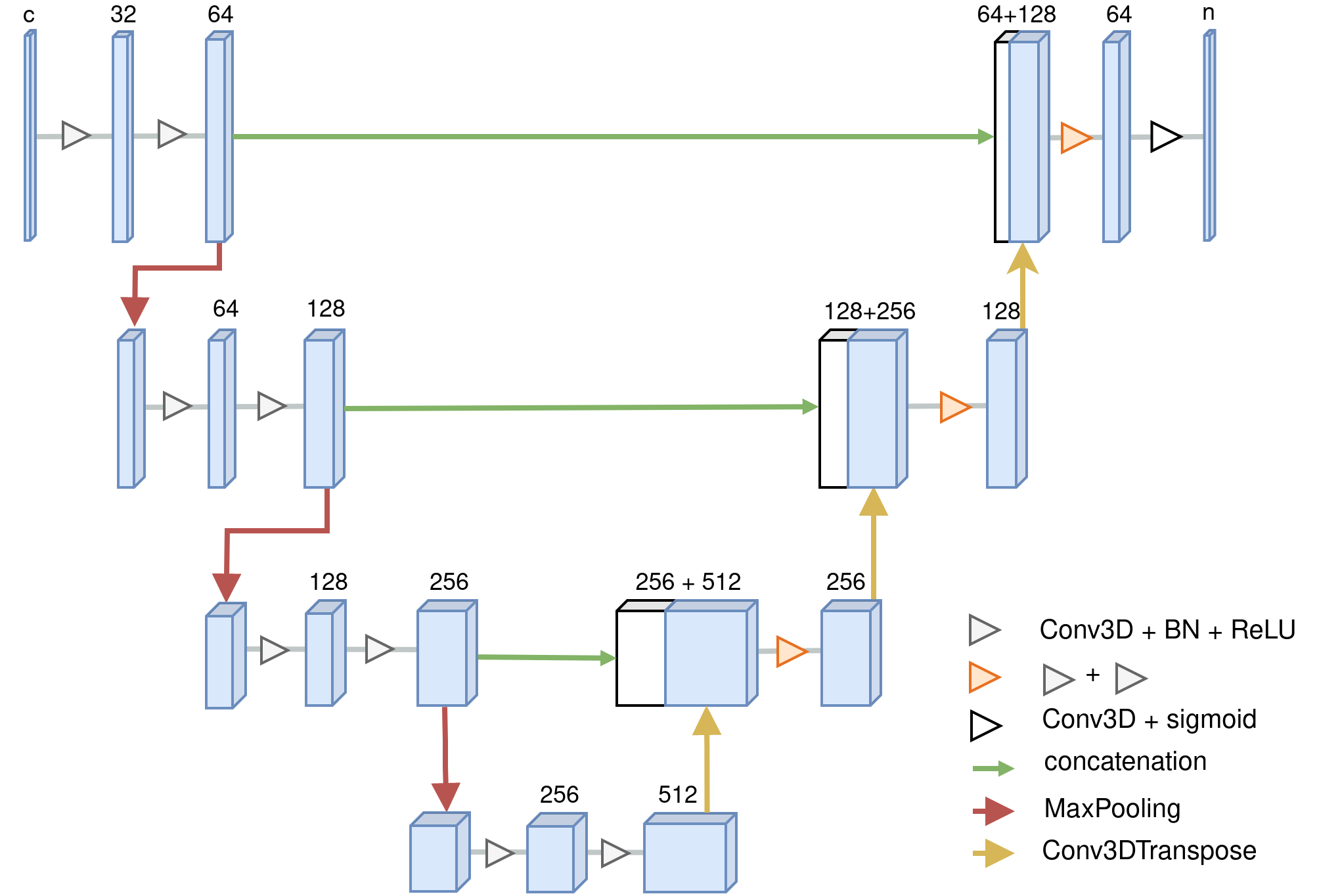
References:
Isensee et al. (2018), Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge, https://arxiv.org/abs/1802.10508
Ronneberger et al. (2015), U-Net: Convolutional Networks for Biomedical Image Segmentation, https://arxiv.org/abs/1505.04597