The Kinetic menu is located in the lower left corner and is mainly used for data operations such as loading, saving or closing. Additionally
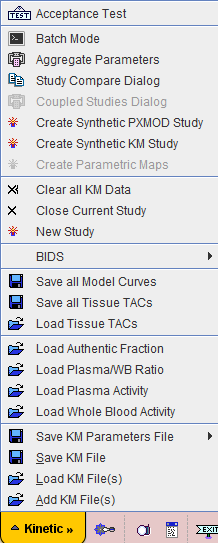
Loaded data sets can be added into new workspaces, so multiple data sets can be available simultaneously and selected for processing using named tabs.
Acceptance Test |
Self test of the program testing 26 model configurations. It is executed automatically the first time PKIN is started. |
Batch Mode |
Open interface for batch processing. |
Aggregate Parameters |
Aggregate the results from different KM Parameter Files (.kinPar) in a single table for statistical analysis. |
Study Compare Dialog |
Open a PKIN dialog window with the same data so that two model configurations can be examined side by side in separate windows. |
Coupled Studies Dialog |
Fit multiple data sets together, keeping selected parameters synchronized. |
Create Synthetic PXMOD Study |
Generate a synthetic image series which has a different combination of model parameters in each pixel. |
Create Synthetic KM Study |
Generate a synthetic modeling data set from the current data with ideal tissue TACs and potentially modified timing. |
Create Parametric Maps |
If the TACs have been transferred from PVIEW to PKIN and the images are still open in PVIEW, open a dialog window for assembling the fitted parameters into images. If average TACs were transferred, the VOIs will be homogeneous showing the regional result. If pixel-wise TACs were transferred, the VOIs will be filled by parametric maps. |
Clear all KM Data |
Close all workspaces. |
Close Current Study |
Close currently selected workspace. |
New Study |
Open a new, empty workspace. |
BIDS |
WIP implementation for creating data sets according to the emerging Bioimaging Data Structure. |
Save all Model Curves |
Save the model curves of all regions in a tab-delimited text file. |
Save all Tissue TACs |
Save the tissue TACs of all regions in a tab-delimited text file. |
Load Tissue TACs |
Load the tissue TACs from a tab-delimited text file. |
Load Authentic Fraction |
Load a fraction curve representing the ratio of parent (aka authentic, intact, unchanged) tracer to total tracer in plasma. |
Load Plasma/WB Ratio |
Load a fraction curve representing the ratio of total tracer in plasma to whole-blood. |
Load Plasma Activity |
Load a curve representing total tracer in plasma. |
Load Whole Blood Activity |
Load a curve representing tracer in whole-blood samples. |
Save KM Parameters File |
Save the parameters of all regional tissue models in a tab-delimited text file (.kinPar). |
Save KM File |
Save all data of the current workspace with all model configurations in the history as a proprietary KM text file (.km). |
Load KM File(s) |
Load a number of selected KM files into separate workspaces, clearing already loaded data. |
Add KM File(s) |
Load a number of selected KM files into additional workspaces, keeping existing data. |