The Cumulated Activity (OLINDA, IDAC) model performs pre-processing steps for the calculation of the absorbed dose from diagnostic or therapeutic radiopharmaceuticals. Given the time-course of the activity in a volume of tissue, it calculates the the Normalized Cumulated Activity [1] (OLINDA Residence Time). The normalized cumulated activity represents the total number of disintegrations which have occurred during an integration time per unit administered activity. Ideally, integration is performed from the time of administration to infinity.
The normalized cumulated activity values resulting from the Cumulated Activity (OLINDA, IDAC) model can serve as input to a program such as OLINDA [2] or IDAC2.1 for the actual calculation of the absorbed organ doses.
Note that all steps required for the analysis of dosimetry data are explained in detail in the PMOD Workflow for Dosimetry Preprocessing application guide.
Operational Equations
Given an activity A0 applied to a subject, and a measured (NOT decay corrected) activity A(t) in an organ, the Normalized Cumulated Activity τ is calculated by
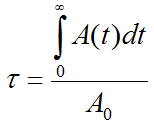
Note that the unit of τ is usually given as [Bq⋅hr/Bq]. The measured part of the organ activity can easily be numerically integrated by the trapezoidal rule. For the unknown remainder of A(t) till infinity it is a conservative assumption to apply the radioactive decay of the isotope. This exponential area can easily be calculated and added to the trapezoidal area. Alternatively, if the measured activity curve has a dominant washout shape, a sum of exponentials can be fitted to the to the measured data and the entire integral algebraically calculated.
The Cumulated Activity (OLINDA, IDAC) model in PKIN supports both the trapezoidal integration approach as well as the use of fitted exponentials. It furthermore supports some practicalities such as
▪conversion of an average activity concentration in [kBq/cc] to VOI activity in [Bq] by multiplication with the VOI volume;
▪reversion of decay correction;
Model Input Parameters
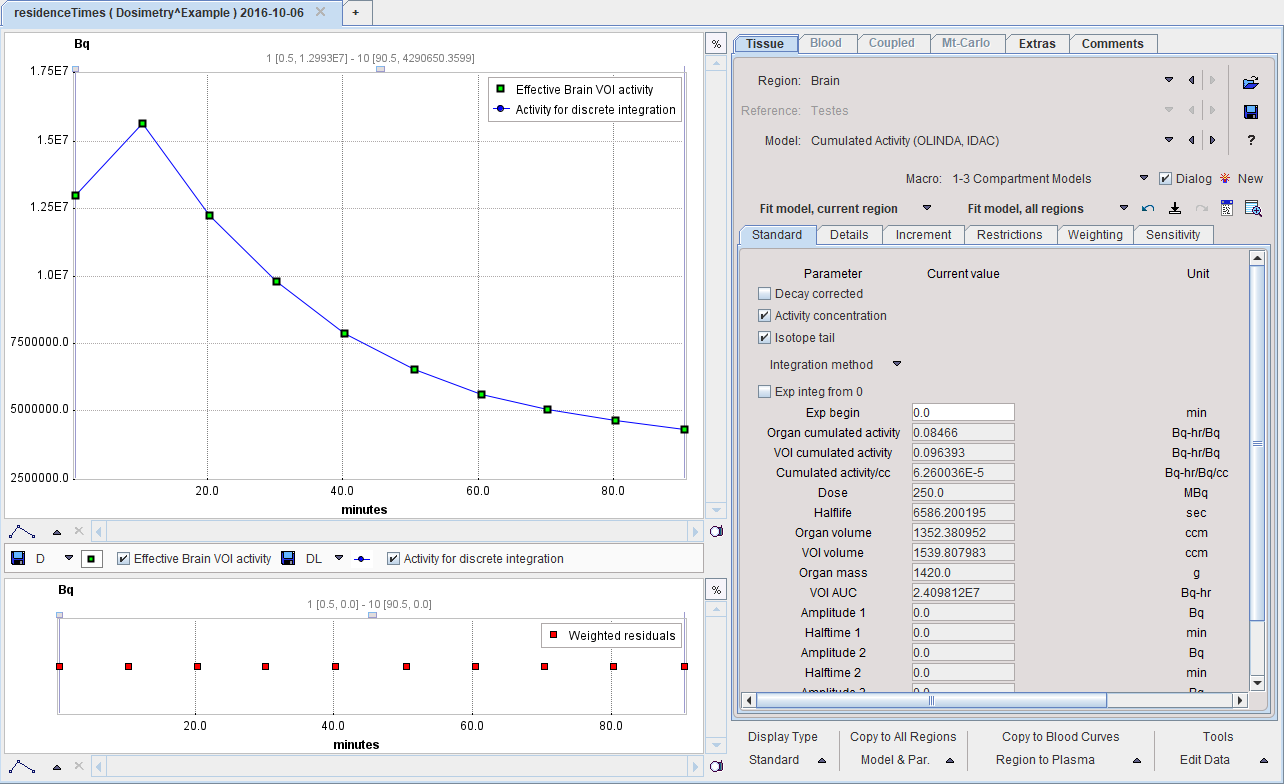
The Cumulated Activity (OLINDA, IDAC) model has 4 input parameters which need to be specified interactively by the user. These settings can easily be propagated from one region to all others with the Model & Par button.
Decay corrected |
Check this box if the loaded curves have been decay corrected as is usually the case for PET imaging studies. The Halflife of the radioisotope will be used to undo the decay correction. |
Activity concentration |
Check this box if the loaded curves represent activity concentration rather than the total activity in a region. The volume of the region will be multiplied with the signal to calculate and show the activity in [Bq]. |
Isotope tail |
Check this box to use the radioactive decay shape for the integration from the end of the last frame to infinity. This option is only relevant if exponentials are fitted to the measurement. For the Rectangle and Trapezoid methods it will always be enabled. For the exponentials it will also be enabled if the exponential halftime is longer than the isotope half-life. |
Integration |
The integration method can be selected in the option list: The Rectangle method is the natural selection for PET values which represent the time average during the frame duration. If there are gaps between the PET frames, the uncovered area is approximated by the trapezoidal rule. The Trapezoid method uses trapezoidal areas between the frame mid-times. With an Exponential selection, the specified number of decaying exponentials is fitted to the measurements, and used for analytical integration. Note that samples marked as invalid are neglected in the fit. |
Exp integ from 0 |
Check this box to use the exponential from time zero for the integration. If the box is unchecked, rectangular integration is used for samples before Exp begin. |
Exp begin |
Defines the time (frame mid), from when on the exponentials are fitted to the measurement. |
Editing of Constituents
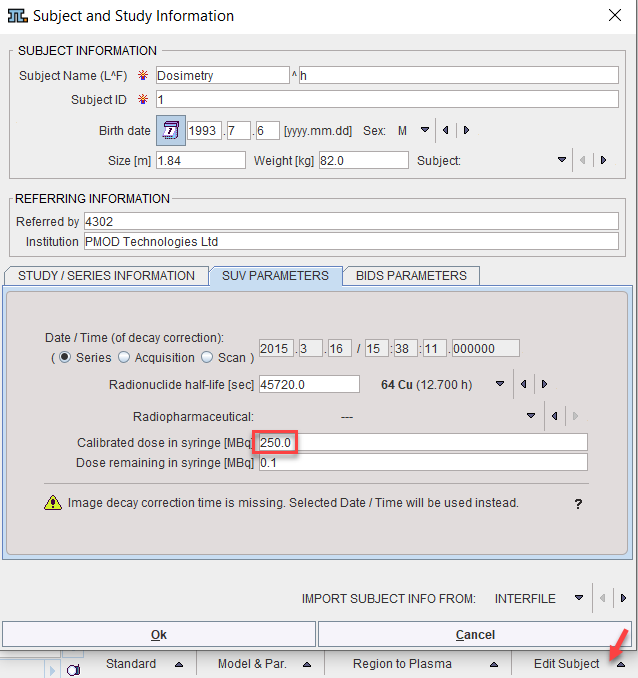
Depending on the import of the activity curves, isotope and activity information may or may not be available. They can be edited in the Subject and Study Information window as illustrated below. Activate Edit Subject in the Tools list, select SUV PARAMETERS, and enter Radionuclide half-life and Calibrated dose in syringe. Note that the activity needs to be calibrated to time 0 of the activity curve.
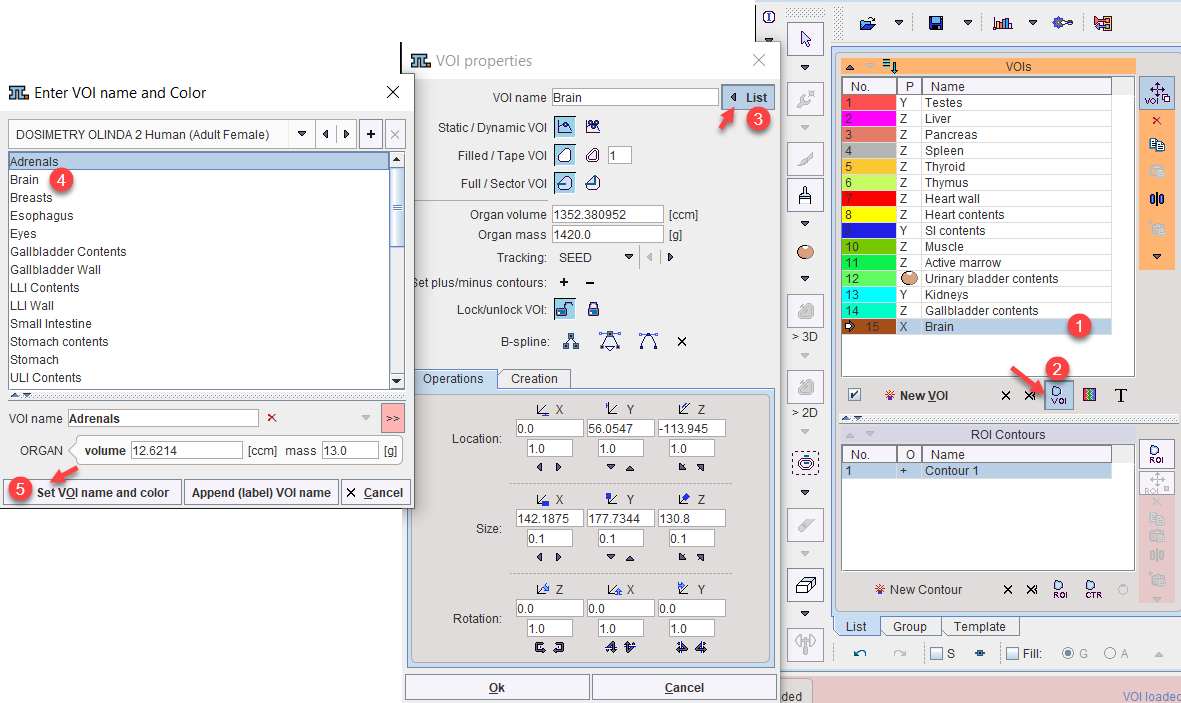
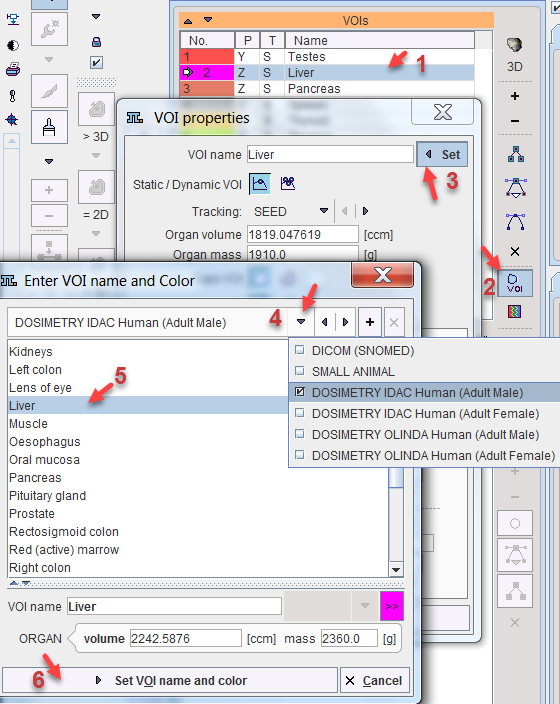
The regional VOI volume is available if the TAC was generated in PMOD's View tool and transferred to PKIN. Often, not the entire volume of the organ can be outlined, because it is not clearly visible, or because (like muscle), it is not compact. In this case, standard organ volumes can be used for extrapolating VOI uptake to organ uptake. An organ can be assigned to a defined VOI as illustrated below:
1.Select the VOI in the list.
2.Open the VOI properties editor, and select the List button.
3.Select an appropriate name list. For instance, the OLINDA 2 Human (Adult Female) contains the organ definitions (name, volume, mass) for the adult male phantom in OLINDA 2.
4.Choose the organ from the list. The predefined ORGAN volume and mass are shown and may be edited.
5.Close with the Set VOI name and color button.
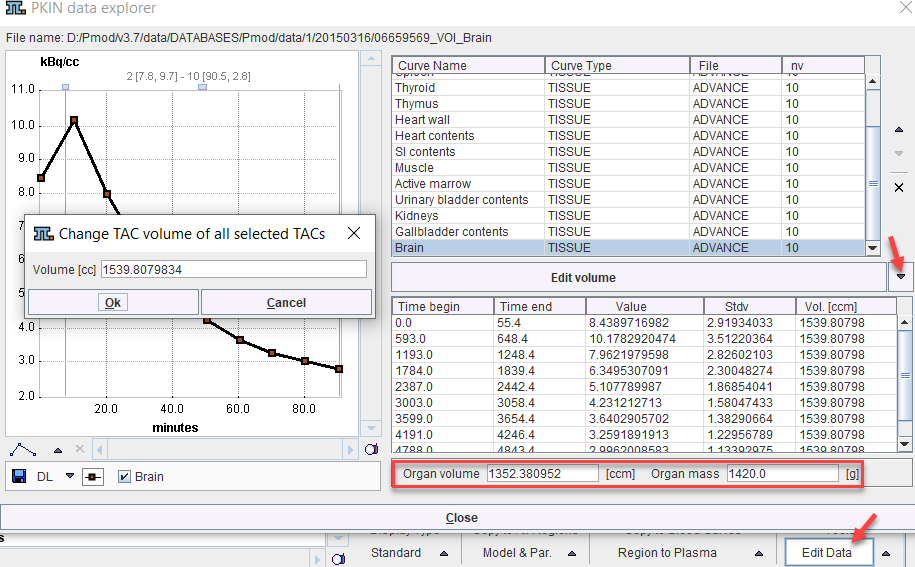
An alternative approach is to edit the VOI and organ volumes as follows:
1.Activate Edit Data in the Tools list
2.Select a region in the upper list.
3.Choose Edit volume from the curve tools
4.Enter the Volume of the VOI in the dialog window.
5.Change The Organ Volume in the lower part. (Note: currently, Organ mass is not used for the calculation).
6.Close the PKIN data explorer with Ok.
Model Output Parameters
As soon as the Fit Region button is activated, the fits and calculations are performed, resulting in a list of parameters:
Organ cumulated activity |
Total number of disintegrations in the organ volume from time 0 until infinity per unit injected activity. This is the main result and equal to Cumulated activity/cc*Organ Volume. Note that it is zero as long as the Organ volume is not defined. |
VOI cumulated activity |
Total number of disintegrations in the regional VOI volume from time 0 until infinity per unit injected activity, calculated as VOIAUC/Activity. |
Cumulated activity/cc |
Total number of disintegrations per unit volume and unit injected activity. Equal to VOI cumulated activity/VOI Volume. |
Dose [MBq] |
Administered activity at the time of scan start. Shown for verification only. Can be changed via Edit Patient. |
Halflife [sec] |
Isotope halflife. Shown for verification only. Can be changed via Edit Patient. |
Organ Volume [ccm] |
Volume of the organ which corresponds to the regional VOI. Shown for verification only. Can be changed via Edit Data. |
Organ Mass [g] |
Volume of the organ which corresponds to the regional VOI. Shown for verification only. Can be changed via Edit Data. |
VOI Volume [ccm] |
Volume of the VOI in which the uptake is measured. Shown for verification only. Can be changed via Edit Data. |
VOI AUC [Bq⋅hr] |
Integral of the VOI activity curve from time 0 till infinity. |
Amplitude 1, 2, 3 [Bq] |
Amplitudes and halftimes of the fitted exponentials. In the case of 1 exponential a linear regression is applied, whereas iterative fitting is used with 2 or 3 exponentials. |
Generate Output File for Dosimetry Programs
After the Cumulated Activity (OLINDA, IDAC) model has been configured and fitted for all organs, a .cas file for direct import into OLINDA or a .idac file for import into IDAC2.1 can be created as follows:
1.Select Edit Data from the Tools list in the lower right.
2.Switch the operation below the curve list to Dosimetry OLINDA/EXM Export or Dosimetry IDAC Export
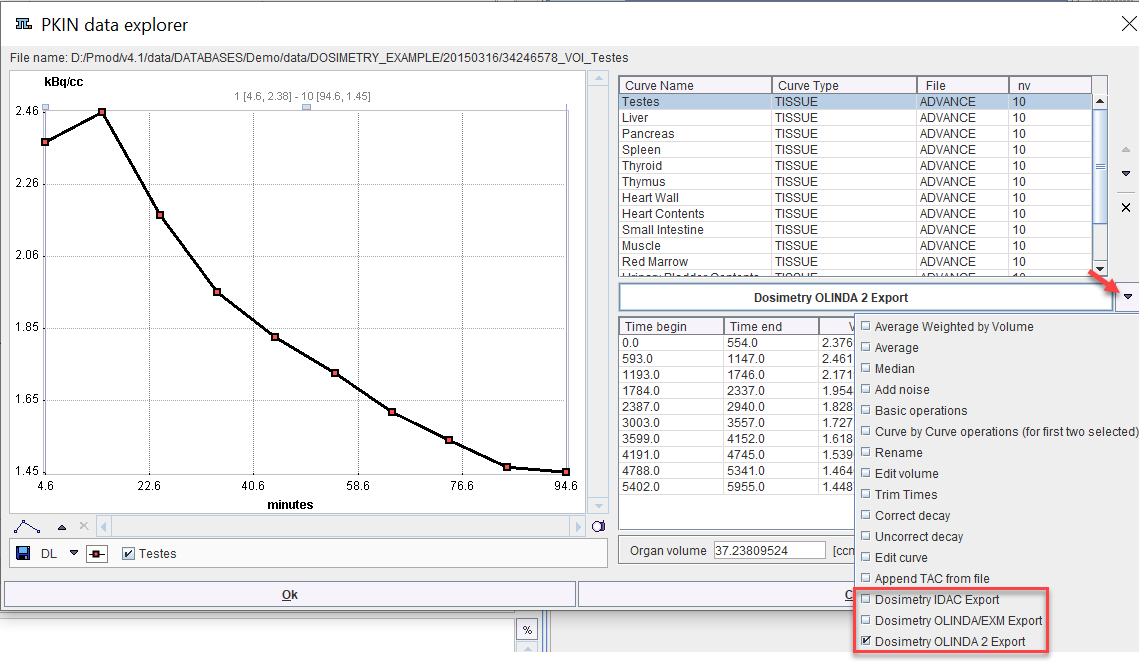
OLINDA/EXM Output
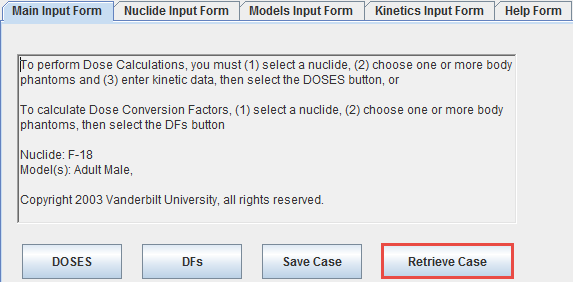
The appearing dialog window
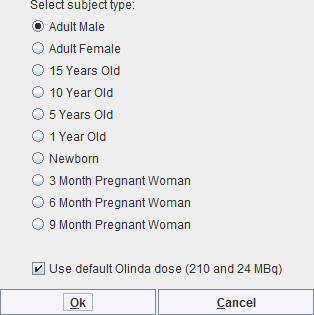
lists the phantoms supported by OLINDA/EXM. Please select the appropriate one, but note that OLINDA/EXM doesn't interpret this information during loading of the .cas file. The option Use default Olinda dose should normally be on. Otherwise, the dose information in the patient SUV panel will be used which will most probably cause problems upon import, because OLINDA/EXM crashes when the dose values are not integer.
Finally specify a location and a name for saving the .cas file which can be loaded in OLINDA/EXM using the Retrieve Case button.
Note that the region names must be spelled exactly as below for the export/import to work properly.
Adrenals
Brain
Breasts_including_skin
Gallbladder_contents
LLI_contents
SI_contents
Stomach_contents
ULI_contents
Heart_contents
Heart_wall
Kidneys
Liver
Lungs
Muscle
Ovaries
Pancreas
Red_marrow
Cortical_bone
Trabecular_bone
Spleen
Testes
Thymus
Thyroid
Urinary_bladder_contents
Uterus
Fetus
Placenta
The difference between the injected activity and the activity accumulated in all organs is treated as the Remainder.
OLINDA 2 Output
The appearing dialog window lists the phantoms supported by the export. Note that within OLINDA 2 the phantoms can be easily switched, whereby the activities are propagated.
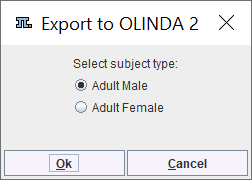
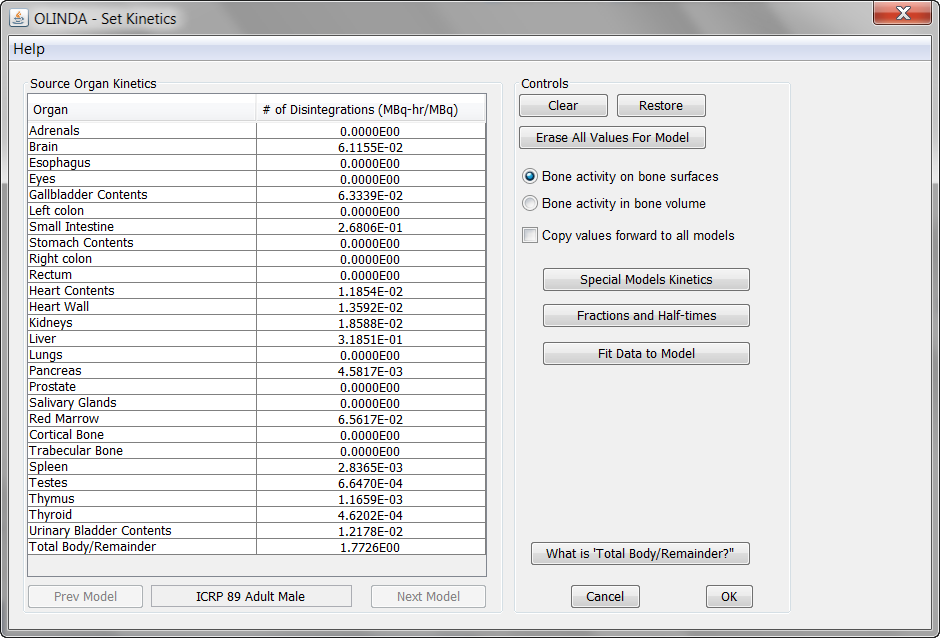
Please select the appropriate one and save the file which is stored with .cas suffix. When opening the file in Olinda 2, the isotope and the model are selected, and the cumulated activities can be inspected on the Set Kinetics panel.
The Remainder value is automatically calculated from the total normalized number of disintegrations of the isotope (=Half-time[h]/ln(2)) minus the activity found in the organs (excluding the bladder). It shows up in the Total Body/Remainder field.
IDAC Output
The appearing dialog window
Please select the appropriate one and save the file which is stored with .idac suffix. When opening the file in IDAC2.1 the cumulated activity will be shown and available for calculating the doses.
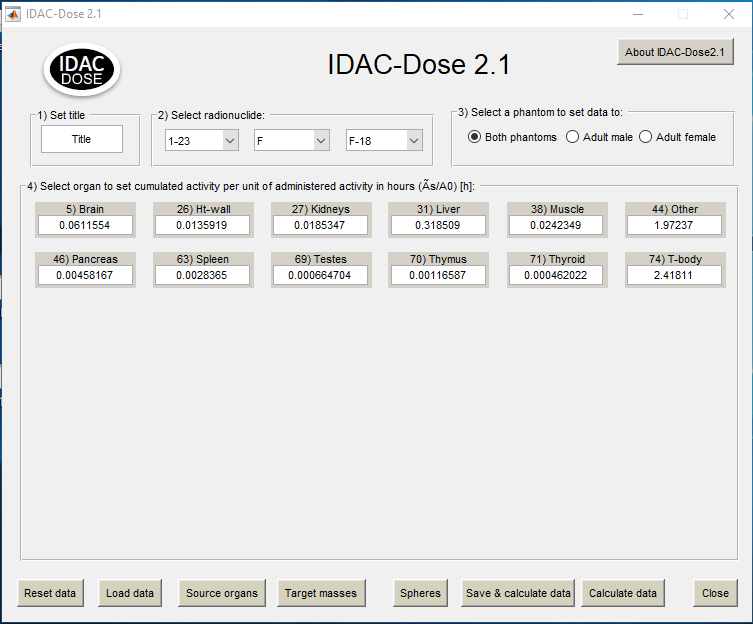
Note that the VOI names for the organs must be defined exactly as follows:
Female |
Male |
Adipose tissue |
Adipose tissue |
Use of Organ Lists for VOI Definition
The OLINDA and IDAC organ lists have been implemented in the VOI facility to support correct VOI naming. Illustrated below is use of these lists to assign proper name and organ mass to a defined VOI.
References
1.Stabin MG: Fundamentals of Nuclear Medicine Dosimetry. Springer; 2008. DOI
2.Stabin MG, Sparks RB, Crowe E: OLINDA/EXM: the second-generation personal computer software for internal dose assessment in nuclear medicine. J Nucl Med 2005, 46(6):1023-1027.